Update: 2021
Avoid CO2 use the Fusion Reactor in the Sky
(if the 2 is pronanced German than it even rhymes)
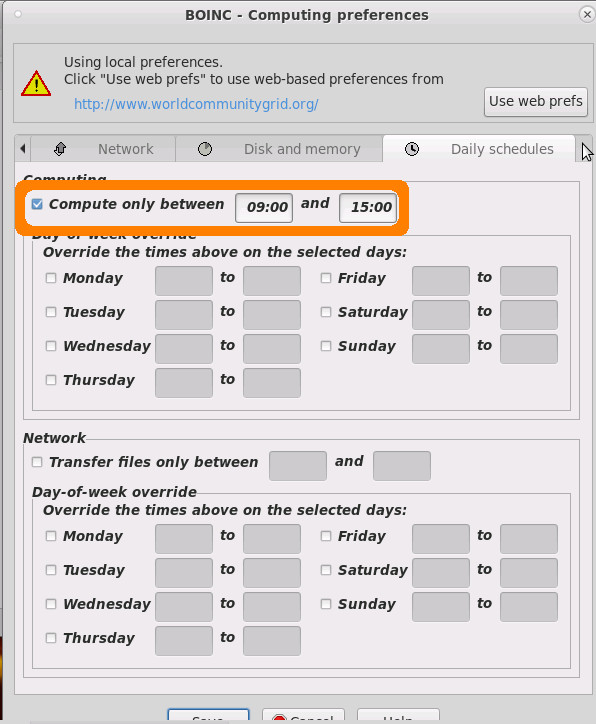
as a lot of fossil fuel is (still in 2021!) used to produce electrictiy, it is possible and recommended to set boinc client to only computer during daytime (when there is the highest %percentage% of solar electrictiy in the grid)
https://www.youtube.com/watch?v=60otR60fPYo
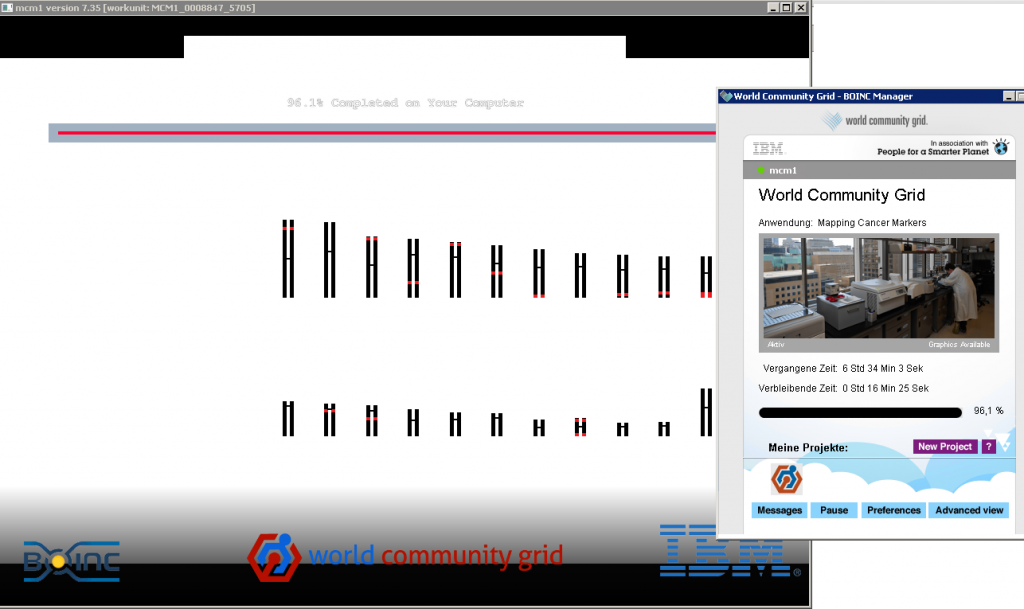
you can find the project at: https://www.worldcommunitygrid.org/research/mcm1/overview.do
- detect cancer earlier
if your server with it’s 48x Xeon CPUs or NVIDIA GPUs (even faster) is idling most of the weekend, why not let it run on this tasks?
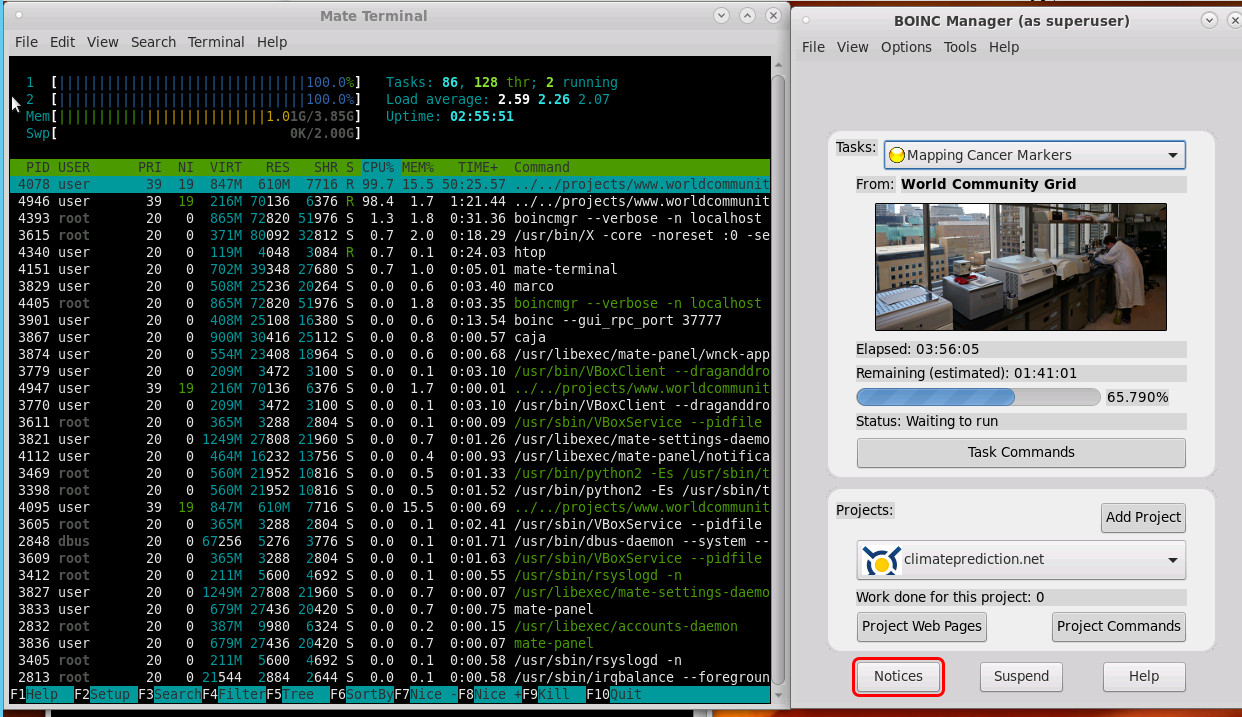
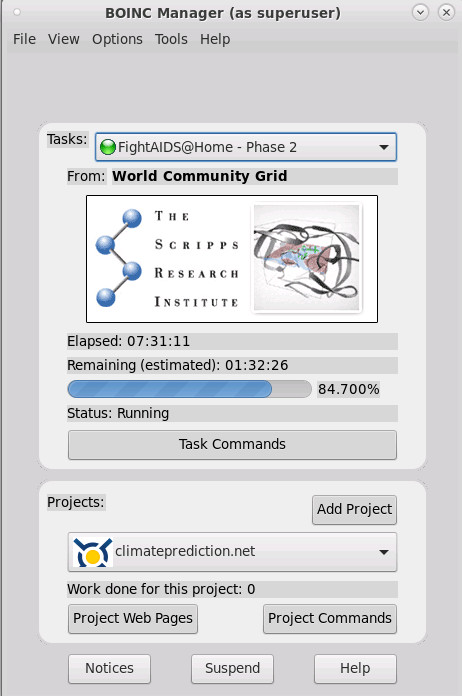
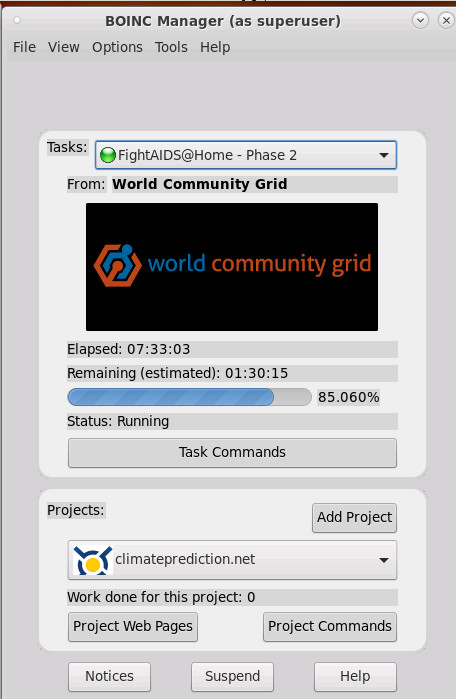
this is what it looks like:
there are loads and loads of projects but also loads and loads of problems with running boinc (unfortunately)
to address (most) of the (current) issues try this: (there is already a rant going on about it, creditz)
# 1. In a terminal window, run the following command: su root apt update apt install boinc-client boinc-manager # 2. Set the BOINC client to automatically start after you restart your computer: systemctl enable boinc-client # 3. Start the BOINC client: systemctl start boinc-client # 4. Allow group access to client access file: chmod g+r /var/lib/boinc-client/gui_rpc_auth.cfg # 5. Add your Linux user to the BOINC group to allow the BOINC Manager to communicate with the BOINC client # use the user's default non-root username usermod -a -G boinc username # problem: to make those changes system wide, reboot might be required X-D # 6. create start script echo 'boincmgr -d /var/lib/boinc-client' > /scripts/boincmgr.sh chmod +x /scripts/*.sh # 7. give it a test run :D /scripts/boincmgr.sh # the boinc client should come up and have no problem connecting to the background boinc processes # one process per cpu core # 8. if that worked, hurray! do a 5min dance of eternal happiness to music of the user's choice # 9. depending on user's desktop add the script to autostart # 10. add or login to a project (WCG is in the list pretty on the bottom)
of course it should not be necessary to run the boinc client (the program that does the actual computing) and the boincmgr as root.
enabling / disabling boinc service under CentOS7 (RedHat/Fedora):
# enable autostart of boinc chkconfig boinc-client on # disable autostart of boinc chkconfig boinc-client off
the boincmgr under CentOS7 (RedHat/Fedora) had some problems connecting to the client process, try this script:
cat /scripts/boinc_start.sh #!/bin/bash export NO_AT_BRIDGE=1; boinc --gui_rpc_port 37777 & boincmgr --verbose -n localhost -g 37777
you can either r-click on the top right corner of your desktop and hit “snooze” if you need the computing power back, or stop/halt the boinc process like this:
service boinc-client stop
- “Cancers, one of the leading causes of death worldwide,
- come in many different types and forms in which uncontrolled cell growth can spread to other parts of the body.
- Unchecked and untreated, cancer can spread from an initial site to other parts of the body and ultimately lead to death.
- The disease is caused by genetic or environmental changes that interfere with biological mechanisms that control cell growth.
- These changes, as well as normal cell activities, can be detected in tissue samples through the presence of their unique chemical indicators, such as DNA and proteins, which together are known as “markers.”
- Specific combinations of these markers may be associated with a given type of cancer.
- The pattern of markers can determine whether an individual is susceptible to developing a specific form of cancer, and may also predict the progression of the disease, helping to suggest the best treatment for a given individual.
- For example, two patients with the same form of cancer may have different outcomes and react differently to the same treatment due to a different genetic profile.
- While several markers are already known to be associated with certain cancers, there are many more to be discovered, as cancer is highly heterogeneous.
- Mapping Cancer Markers on World Community Grid aims to identify the markers associated with various types of cancer.
- The project is analyzing millions of data points collected from thousands of healthy and cancerous patient tissue samples.
- These include tissues with lung, ovarian, prostate, pancreatic and breast cancers.
- By comparing these different data points, researchers aim to identify patterns of markers for different cancers and correlate them with different outcomes, including responsiveness to various treatment options.
- This knowledge can help researchers and physicians to:
- Improve and personalize cancer treatment
- by making it possible to detect cancer earlier
- identify high-risk patients
- and customize treatment based on a patient’s personal genetic profile
- Accelerate cancer research and improve the overall process for identifying markers
- by refining the process of identifying markers, researchers can determine an individual patient’s markers more easily, and future large sets of data can be analyzed more efficiently.
- Improve and personalize cancer treatment
- src: https://www.worldcommunitygrid.org/research/mcm1/overview.do
“Cancer development is a multi-step process that leads to uncontrolled tumour cell growth caused by and resulting in complex changes: many genes are amplified, deleted, mutated, up- or down-regulated; many proteins and pathways are activated or suppressed. Estimating across 1.9 million patients from 31 countries and 5 continents, current treatments achieve a 5-year survival rate for less than 50% of diagnosed cancer (Coleman et al. Cancer survival in five continents: a worldwide population-based study (CONCORD).
Lancet Oncol 9(8): 730-756, 2008).
Years of research improved survival in breast and prostate cancers by finding molecular markers for early diagnosis and by individualized treatment. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
The Mapping Cancer Markers (MCM) project aims to comprehensively and systematically discover clinically useful markers to aid early cancer detection, identification of high-risk patients, and prediction of treatment response.
To power this research, we rely on World Community Grid volunteers who donate their computers’ spare capacity to carry out this extensive analysis. Finding all clinically useful markers would require processing thousands of patient samples and testing an astronomical number of marker combinations, which is not feasible even on World Community Grid. Instead, we use heuristics to reduce the search space, enabling us to tackle this challenge with the computing resources donated by volunteers like you.
Support our research and join World Community Grid today!”
src: https://www.cs.utoronto.ca/~juris/MCM.htm
Cancer Challenge
Cancer development is a multi-step process that leads to uncontrolled tumour cell growth caused by and resulting in complex changes: many genes are amplified, deleted, mutated, up- or down-regulated, many pathways are activated or suppressed.Estimating from the CONCORD study across 1.9 million patients from 31 countries and 5 continents, current treatments achieve a 5-year survival rate for less than 50% of diagnosed cancer (Coleman, 2008). Approximately 12.7 million new cancer cases were diagnosed worldwide in 2008. Lung, female breast, colorectal & stomach cancers account for more than 40% of all cases. Almost 8 million deaths from cancer occurred worldwide in 2008. Lung, stomach, liver, colorectal & female breast cancers account for more than 50% of all cancer deaths (http://www.cancerresearchuk.org/cancer-info/cancerstats/world/).
Canadian cancer statistics (2012) indicates that lung cancer accounts for almost 14% of all cancer cases in Canada, leading to the highest number of deaths (20,100, 27% of all cancers). Years of research improved relative survival by 6% for all cancers since 1992. Highest improvements are for non-Hodgkin lymphoma & leukemias. Success in breast and prostate cancers has been achieved by finding molecular markers for early diagnosis and by individualized treatment. Highest survival is for thyroid, prostate & testicular cancers. However, pancreatic cancer remains almost 100% lethal, and the overall survival rate for lung cancer has improved barely during the past decades, having only moved from 13% to 16%.
Cancer is caused by genetic changes or environmental effects that interfere with the mechanisms that control cell growth. These changes, as well as normal cell activities, can be detected in tissue samples through the presence of unique indicators, such as DNA and proteins, which together are known as “markers.” Specific combinations of these markers may be associated with a given type of cancer, indicate patient’s risk of cancer recurrence, or indicate probability of patient’s response to treatment. While several markers are already known to be associated with certain cancers, there are many more to be discovered, as cancer is highly heterogeneous.
The discovery and validation of biomarkers is complex and computationally intensive process. It involves analyzing hundreds of thousands of parameters (clinical variables, gene, protein, microRNA, activity, etc.) to identify subsets that best describe patients, their prognosis and response to treatment. Finding all clinically useful markers and selecting the best subset represents a challenging computational optimization task as we would need to compare all possible parameter combinations.
Approach
The Mapping Cancer Markers (MCM) project focuses on clinical application – discovering specific groups of markers that can be used to improve detection, diagnosis, prognosis and treatment of cancer. As a second goal, the comprehensive analysis of existing molecular profiles of cancer samples will lead to unraveling characteristics of such groups of markers – and in turn improving our understanding how to find them more efficiently.
Our strategy to reduce mortality includes three steps:
- Increase number of cases diagnosed at earlier stage
- We need to identify biomarkers for early cancer detection
- Individualized treatment
- We need to find biomarkers for treatment selection & response monitoring
- Improved treatment
-
- We need to improve our understanding of disease mechanism and drug mechanism_of_action
- We need to identify useful drug combinations & design new medicines
To address these challenges, we propose to conduct an integrative analysis and comprehensive computational and biological validation of putative markers. To find all good markers, requires to use large patient cohorts (thousands of patient samples) and test billions of marker combinations, which would be intractable even on the World Community Grid. Thus, we have developed software which uses heuristics (clever steps that reduce the enormous search space by focusing the search on the most relevant subsets of combinations) which can greatly reduce the computational effort required to look for significant marker patterns. Even these software methods require a very large amount of computer processing power. By using World Community Grid, the Mapping Cancer Markers researchers are able to break down this overwhelming process into smaller, manageable tasks which can be performed by our volunteers’ computing devices. World Community Grid therefore allows researchers to undertake this ambitious research. Thus, MCM focuses on three goals:
- Identify sets of markers that may be able to predict if a person is at high risk of developing a particular cancer and increase the possibility of early detection.
- Identify combinations of markers, which may predict a patient’s response to specific treatments. This would help make the treatment more personalized and could guide the development of customized therapeutic treatments for that patient.
- Develop more efficient computational methods for discovering relevant patterns of markers.
References
Coleman, M. P., M. Quaresma, et al. (2008). “Cancer survival in five continents: a worldwide population-based study (CONCORD).” Lancet Oncol 9(8): 730-756.
Ein-Dor, L., I. Kela, et al. (2005). “Outcome signature genes in breast cancer: is there a unique set?” Bioinformatics 21(2): 171-178.
Ein-Dor, L., O. Zuk, et al. (2006). “Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer.” Proc Natl Acad Sci U S A 103(15): 5923-5928.
src: https://www.cs.utoronto.ca/~juris/MCMpd.html
Links:
https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer
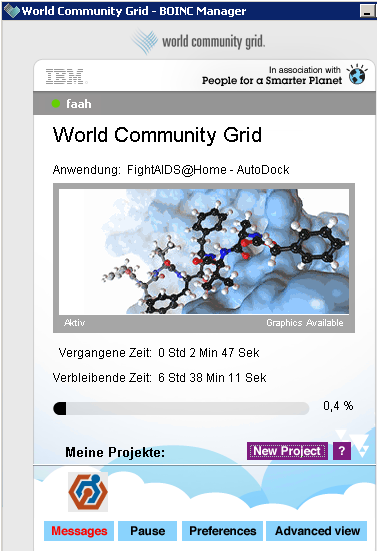
WCG is also Fighting AIDS
World Community Grid is a distributed computing project by IBM allowing scientists to run digital experiments on their infrastructure. It is based on the BOINC client by University of Berkeley and available for Windows, Linux and OSX.
Its an effort to create the world’s largest public computing grid to tackle scientific research projects that benefit humanity.[4] Launched on November 16, 2004, it is co-ordinated by IBM.
If you can spare some kWh I recommend to only run it on proper servers (Xeon) or maybe self-build NAS with massive spare CPU / GPU powers that are operating 24-7 anythow and sit idle on sundays and night times, you could run it on a rented VPS / your webserver but it will eat like 512MB of RAM.
search for extraterrestrial intelligence (SETI) is using it too https://seti.org/
So no client-hard or software, not laptops, no workstations – while it tries to not use up all your ressources it will slow down your client.
If you want to setup an account, feel free to use my invitation link 🙂
You can also hook it up to your twitter account and it will post messages like once a month like:
I have contributed 6 years to humanitarian scientific research through @WCGrid. Join me: https://autotweet.worldcommunitygrid.org?recruiterId=265744autotweet.worldcommunitygrid.org/?recruiterId=2 …
IBM needs to make sure that this massive computing power is never used for evil.
They used to have nice graphical display of what project is currently under work on your computer.
on a 8x Core Xeon Server with 2.0 Ghz i let this app run with 50% of one Core 24-7.
Praying for a better world. With less cancer.
You can get an account here: http://www.worldcommunitygrid.org/
Download the software here: http://www.worldcommunitygrid.org/clients/wcg_boinc_7.2.47_windows_intelx86.exe
It also Supports some special GPUs with PhysX which should have much better MFLops/Watt ratio.
more on the command line:
Linux (in general)
The “init” script which starts or stops the daemon is the same on any Linux distribution (including Ubuntu or Debian, or Fedora or Red Hat, or SUSE). It is used by the system at boot time or shutdown time to start or stop the daemon. You can use it directly from the command line to start or stop the daemon, or check the status. You must be root to run this script, so you must either login as root, use the ‘sudo’ command, or ‘su’ to root.
/etc/init.d/boinc-client start- – start BOINC client, running as a system daemon
/etc/init.d/boinc-client stop- – stop the BOINC daemon
/etc/init.d/boinc-client restart- – stop and start the BOINC daemon
/etc/init.d/boinc-client status- – report on status of the daemon (is it running or not?)
Fedora/Red Hat
Using the command line
In Fedora and Red Hat distributions and those based on Red Hat (like CentOS or Mandrake) you use the chkconfig command to turn a service (daemon) on or off at boot time. In addition to running the “init” script directly, as shown above, you can also use the service command to start or stop a daemon. Remember that you must be root to run these commands.
chkconfig boinc-client off- -tells Fedora to not start the daemon at boot time (but does not stop a running daemon)
chkconfig boinc-client on- -tells Fedora to start the daemon at boot time (but does not start the daemon immediately)
service boinc-client stop- -stops a running daemon
service boinc-client start- -starts the daemon
service boinc-client restart- -stops and then restarts the daemon
If you use sudo to run these commands you will need to give the full-path name of the command, which means /sbin/chkconfig or /sbin/service.
Using the GUI
The services GUI provides an easy way to turn the boinc-client service on or off (have it start or not start at boot time), start or stop the service after you have booted or just check if the service is running.
On Fedora 5 with KDE, click Applications -> Administration -> Server Settings -> Services. On Fedora 9 with KDE, click Applications -> Administration -> Service Management. Fedora 6, 7, 8 and 10 should be fairly similar to either Fedora 5 or 9.
Ubuntu/Debian
In Debian-based Linux distributions you use the update-rc.d command to turn a system service (daemon) on or off at boot time.
update-rc.d boinc-client defaults 98- -tells the system to start the BOINC client as a daemon at boot time
update-rc.d boinc-client remove- – tells the system not to start the BOINC client at boot time
See the man page for update-rc.d for more details.
Gentoo
In Gentoo-based Linux distributions you use the rc-update command to turn a system service (daemon) on or off at boot time.
rc-update add boinc-client default- -tells the system to start the BOINC client as a daemon at boot time (after networking support has been enabled)
rc-update del boinc-client- – tells the system not to start the BOINC client at boot time
See the man page for rc-update for more details.
SUSE Linux
In opensuse Linux distributions you use the insserv command to turn a system service (daemon) on or off at boot time.
insserv boinc-client- – tells the system to start the BOINC client as a daemon at boot time
insserv -r boinc-client- – removes the BOINC client from the list of services started at boot time
An init script for opensuse is available.
src: https://boinc.berkeley.edu/wiki/Stop_or_start_BOINC_daemon_after_boot
News:
https://twitter.com/hashtag/wcg
https://twitter.com/hashtag/boinc
https://twitter.com/hashtag/seti
liked this article?
- only together we can create a truly free world
- plz support dwaves to keep it up & running!
- (yes the info on the internet is (mostly) free but beer is still not free (still have to work on that))
- really really hate advertisement
- contribute: whenever a solution was found, blog about it for others to find!
- talk about, recommend & link to this blog and articles
- thanks to all who contribute!